doi.org/10.1038/srep42362
Preview meta tags from the doi.org website.
Linked Hostnames
36- 179 links todoi.org
- 43 links toscholar.google.com
- 28 links towww.ncbi.nlm.nih.gov
- 23 links towww.nature.com
- 7 links towww.springernature.com
- 4 links topartnerships.nature.com
- 4 links toscholar.google.co.uk
- 3 links toauthorservices.springernature.com
Thumbnail
Search Engine Appearance
Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC - Scientific Reports
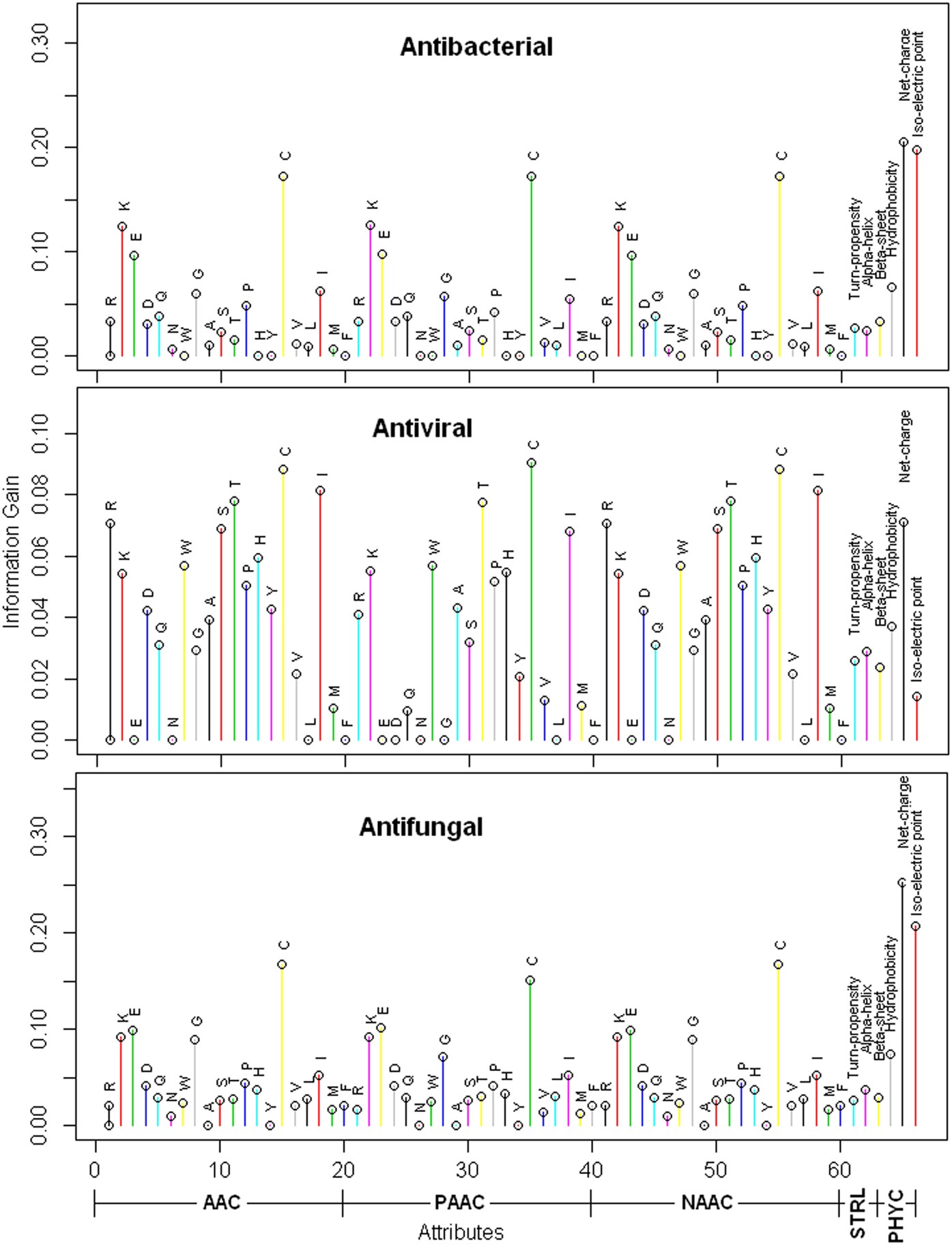
Antimicrobial peptides (AMPs) are important components of the innate immune system that have been found to be effective against disease causing pathogens. Identification of AMPs through wet-lab experiment is expensive. Therefore, development of efficient computational tool is essential to identify the best candidate AMP prior to the in vitro experimentation. In this study, we made an attempt to develop a support vector machine (SVM) based computational approach for prediction of AMPs with improved accuracy. Initially, compositional, physico-chemical and structural features of the peptides were generated that were subsequently used as input in SVM for prediction of AMPs. The proposed approach achieved higher accuracy than several existing approaches, while compared using benchmark dataset. Based on the proposed approach, an online prediction server iAMPpred has also been developed to help the scientific community in predicting AMPs, which is freely accessible at http://cabgrid.res.in:8080/amppred/. The proposed approach is believed to supplement the tools and techniques that have been developed in the past for prediction of AMPs.
Bing
Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC - Scientific Reports
Antimicrobial peptides (AMPs) are important components of the innate immune system that have been found to be effective against disease causing pathogens. Identification of AMPs through wet-lab experiment is expensive. Therefore, development of efficient computational tool is essential to identify the best candidate AMP prior to the in vitro experimentation. In this study, we made an attempt to develop a support vector machine (SVM) based computational approach for prediction of AMPs with improved accuracy. Initially, compositional, physico-chemical and structural features of the peptides were generated that were subsequently used as input in SVM for prediction of AMPs. The proposed approach achieved higher accuracy than several existing approaches, while compared using benchmark dataset. Based on the proposed approach, an online prediction server iAMPpred has also been developed to help the scientific community in predicting AMPs, which is freely accessible at http://cabgrid.res.in:8080/amppred/. The proposed approach is believed to supplement the tools and techniques that have been developed in the past for prediction of AMPs.
DuckDuckGo
Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC - Scientific Reports
Antimicrobial peptides (AMPs) are important components of the innate immune system that have been found to be effective against disease causing pathogens. Identification of AMPs through wet-lab experiment is expensive. Therefore, development of efficient computational tool is essential to identify the best candidate AMP prior to the in vitro experimentation. In this study, we made an attempt to develop a support vector machine (SVM) based computational approach for prediction of AMPs with improved accuracy. Initially, compositional, physico-chemical and structural features of the peptides were generated that were subsequently used as input in SVM for prediction of AMPs. The proposed approach achieved higher accuracy than several existing approaches, while compared using benchmark dataset. Based on the proposed approach, an online prediction server iAMPpred has also been developed to help the scientific community in predicting AMPs, which is freely accessible at http://cabgrid.res.in:8080/amppred/. The proposed approach is believed to supplement the tools and techniques that have been developed in the past for prediction of AMPs.
General Meta Tags
125- titlePredicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC | Scientific Reports
- titleClose banner
- titleClose banner
- X-UA-CompatibleIE=edge
- applicable-devicepc,mobile
Open Graph Meta Tags
5- og:urlhttps://www.nature.com/articles/srep42362
- og:typearticle
- og:site_nameNature
- og:titlePredicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC - Scientific Reports
- og:imagehttps://media.springernature.com/m685/springer-static/image/art%3A10.1038%2Fsrep42362/MediaObjects/41598_2017_Article_BFsrep42362_Fig1_HTML.jpg
Twitter Meta Tags
6- twitter:site@SciReports
- twitter:cardsummary_large_image
- twitter:image:altContent cover image
- twitter:titlePredicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC
- twitter:descriptionScientific Reports - Predicting antimicrobial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC
Item Prop Meta Tags
5- position1
- position2
- position3
- position4
- publisherSpringer Nature
Link Tags
15- alternatehttps://www.nature.com/srep.rss
- apple-touch-icon/static/images/favicons/nature/apple-touch-icon-f39cb19454.png
- canonicalhttps://www.nature.com/articles/srep42362
- icon/static/images/favicons/nature/favicon-48x48-b52890008c.png
- icon/static/images/favicons/nature/favicon-32x32-3fe59ece92.png
Emails
1Links
324- http://adsabs.harvard.edu/cgi-bin/nph-data_query?link_type=ABSTRACT&bibcode=2011PLoSO...618476W
- http://adsabs.harvard.edu/cgi-bin/nph-data_query?link_type=ABSTRACT&bibcode=2013PLoSO...866557Z
- http://cabgrid.res.in:8080/amppred
- http://cabgrid.res.in:8080/amppred/about.html
- http://crdd.osdd.net/servers/avppred/collection.php?show=dataset