doi.org/10.1038/ncomms8822
Preview meta tags from the doi.org website.
Linked Hostnames
32- 249 links todoi.org
- 65 links toscholar.google.com
- 45 links towww.ncbi.nlm.nih.gov
- 33 links toscholar.google.co.uk
- 22 links towww.nature.com
- 16 links toadsabs.harvard.edu
- 11 links tolink.springer.com
- 7 links towww.springernature.com
Thumbnail
Search Engine Appearance
The iBeetle large-scale RNAi screen reveals gene functions for insect development and physiology - Nature Communications
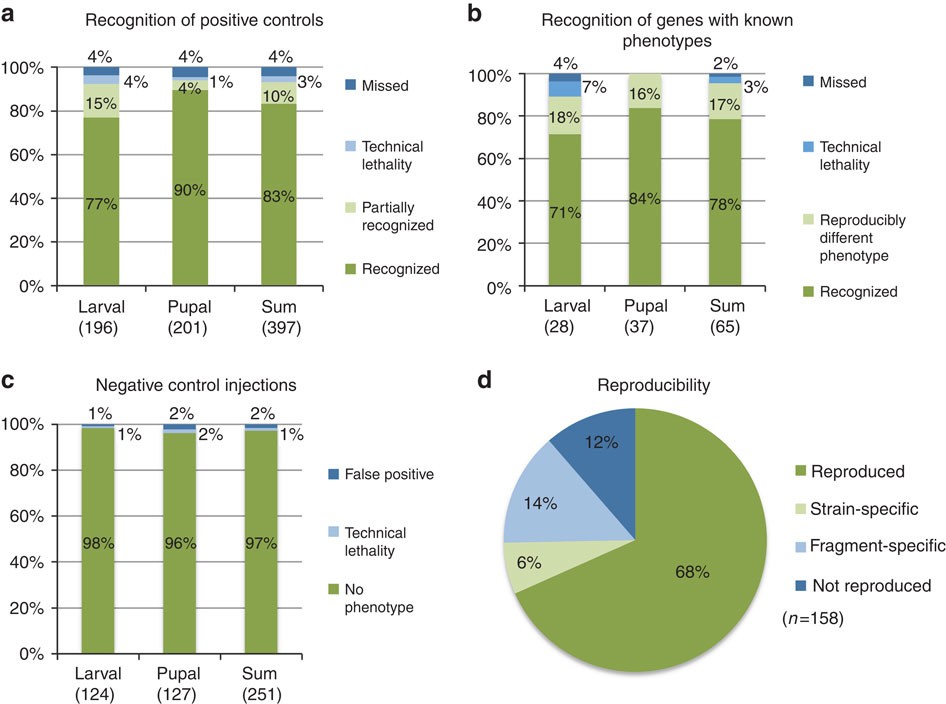
Genetic screens are powerful tools to identify the genes required for a given biological process. However, for technical reasons, comprehensive screens have been restricted to very few model organisms. Therefore, although deep sequencing is revealing the genes of ever more insect species, the functional studies predominantly focus on candidate genes previously identified in Drosophila, which is biasing research towards conserved gene functions. RNAi screens in other organisms promise to reduce this bias. Here we present the results of the iBeetle screen, a large-scale, unbiased RNAi screen in the red flour beetle, Tribolium castaneum, which identifies gene functions in embryonic and postembryonic development, physiology and cell biology. The utility of Tribolium as a screening platform is demonstrated by the identification of genes involved in insect epithelial adhesion. This work transcends the restrictions of the candidate gene approach and opens fields of research not accessible in Drosophila. Unbiased screening for insect gene function has been largely restricted to Drosophila. Here, Schmitt-Engel et al. perform an unbiased large-scale RNAi screen in the red flour beetle Tribolium castaneumto identify putative gene functions.
Bing
The iBeetle large-scale RNAi screen reveals gene functions for insect development and physiology - Nature Communications
Genetic screens are powerful tools to identify the genes required for a given biological process. However, for technical reasons, comprehensive screens have been restricted to very few model organisms. Therefore, although deep sequencing is revealing the genes of ever more insect species, the functional studies predominantly focus on candidate genes previously identified in Drosophila, which is biasing research towards conserved gene functions. RNAi screens in other organisms promise to reduce this bias. Here we present the results of the iBeetle screen, a large-scale, unbiased RNAi screen in the red flour beetle, Tribolium castaneum, which identifies gene functions in embryonic and postembryonic development, physiology and cell biology. The utility of Tribolium as a screening platform is demonstrated by the identification of genes involved in insect epithelial adhesion. This work transcends the restrictions of the candidate gene approach and opens fields of research not accessible in Drosophila. Unbiased screening for insect gene function has been largely restricted to Drosophila. Here, Schmitt-Engel et al. perform an unbiased large-scale RNAi screen in the red flour beetle Tribolium castaneumto identify putative gene functions.
DuckDuckGo
The iBeetle large-scale RNAi screen reveals gene functions for insect development and physiology - Nature Communications
Genetic screens are powerful tools to identify the genes required for a given biological process. However, for technical reasons, comprehensive screens have been restricted to very few model organisms. Therefore, although deep sequencing is revealing the genes of ever more insect species, the functional studies predominantly focus on candidate genes previously identified in Drosophila, which is biasing research towards conserved gene functions. RNAi screens in other organisms promise to reduce this bias. Here we present the results of the iBeetle screen, a large-scale, unbiased RNAi screen in the red flour beetle, Tribolium castaneum, which identifies gene functions in embryonic and postembryonic development, physiology and cell biology. The utility of Tribolium as a screening platform is demonstrated by the identification of genes involved in insect epithelial adhesion. This work transcends the restrictions of the candidate gene approach and opens fields of research not accessible in Drosophila. Unbiased screening for insect gene function has been largely restricted to Drosophila. Here, Schmitt-Engel et al. perform an unbiased large-scale RNAi screen in the red flour beetle Tribolium castaneumto identify putative gene functions.
General Meta Tags
232- titleThe iBeetle large-scale RNAi screen reveals gene functions for insect development and physiology | Nature Communications
- titleClose banner
- titleClose banner
- X-UA-CompatibleIE=edge
- applicable-devicepc,mobile
Open Graph Meta Tags
6- og:urlhttps://www.nature.com/articles/ncomms8822
- og:typearticle
- og:site_nameNature
- og:titleThe iBeetle large-scale RNAi screen reveals gene functions for insect development and physiology - Nature Communications
- og:descriptionUnbiased screening for insect gene function has been largely restricted to Drosophila. Here, Schmitt-Engel et al. perform an unbiased large-scale RNAi screen in the red flour beetle Tribolium castaneumto identify putative gene functions.
Twitter Meta Tags
6- twitter:site@NatureComms
- twitter:cardsummary_large_image
- twitter:image:altContent cover image
- twitter:titleThe iBeetle large-scale RNAi screen reveals gene functions for insect development and physiology
- twitter:descriptionNature Communications - Unbiased screening for insect gene function has been largely restricted to Drosophila. Here, Schmitt-Engel et al. perform an unbiased large-scale RNAi screen in the red...
Item Prop Meta Tags
5- position1
- position2
- position3
- position4
- publisherSpringer Nature
Link Tags
15- alternatehttps://www.nature.com/ncomms.rss
- apple-touch-icon/static/images/favicons/nature/apple-touch-icon-f39cb19454.png
- canonicalhttps://www.nature.com/articles/ncomms8822
- icon/static/images/favicons/nature/favicon-48x48-b52890008c.png
- icon/static/images/favicons/nature/favicon-32x32-3fe59ece92.png
Emails
2Links
482- http://adsabs.harvard.edu/cgi-bin/nph-data_query?link_type=ABSTRACT&bibcode=1980Natur.287..795N
- http://adsabs.harvard.edu/cgi-bin/nph-data_query?link_type=ABSTRACT&bibcode=1994Sci...266..572N
- http://adsabs.harvard.edu/cgi-bin/nph-data_query?link_type=ABSTRACT&bibcode=1999Natur.402..370B
- http://adsabs.harvard.edu/cgi-bin/nph-data_query?link_type=ABSTRACT&bibcode=2000Natur.408..325F
- http://adsabs.harvard.edu/cgi-bin/nph-data_query?link_type=ABSTRACT&bibcode=2000Natur.408..331G