doi.org/10.7717/peerj-cs.61
Preview meta tags from the doi.org website.
Linked Hostnames
23- 51 links todoi.org
- 23 links todfzljdn9uc3pi.cloudfront.net
- 4 links topeerj.com
- 3 links tobioassayontology.org
- 3 links towww.facebook.com
- 2 links tobioportal.bioontology.org
- 2 links togithub.com
- 2 links totwitter.com
Thumbnail
Search Engine Appearance
BioAssay templates for the semantic web
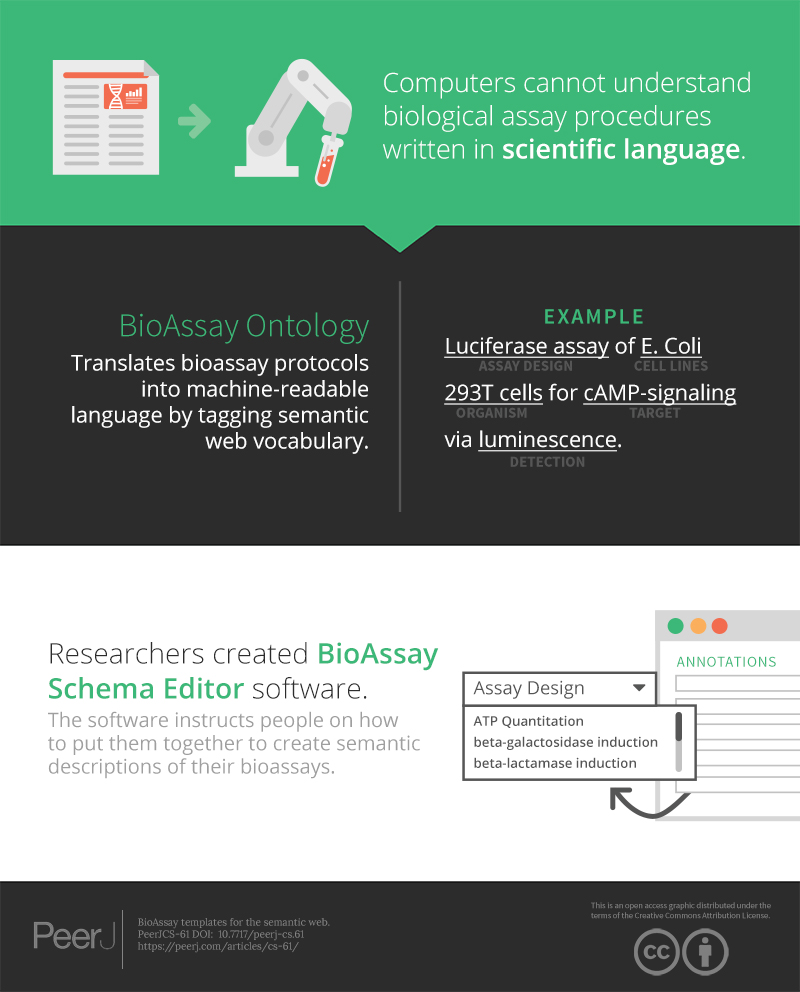
Annotation of bioassay protocols using semantic web vocabulary is a way to make experiment descriptions machine-readable. Protocols are communicated using concise scientific English, which precludes most kinds of analysis by software algorithms. Given the availability of a sufficiently expressive ontology, some or all of the pertinent information can be captured by asserting a series of facts, expressed as semantic web triples (subject, predicate, object). With appropriate annotation, assays can be searched, clustered, tagged and evaluated in a multitude of ways, analogous to other segments of drug discovery informatics. The BioAssay Ontology (BAO) has been previously designed for this express purpose, and provides a layered hierarchy of meaningful terms which can be linked to. Currently the biggest challenge is the issue of content creation: scientists cannot be expected to use the BAO effectively without having access to software tools that make it straightforward to use the vocabulary in a canonical way. We have sought to remove this barrier by: (1) defining a BioAssay Template (BAT) data model; (2) creating a software tool for experts to create or modify templates to suit their needs; and (3) designing a common assay template (CAT) to leverage the most value from the BAO terms. The CAT was carefully assembled by biologists in order to find a balance between the maximum amount of information captured vs. low degrees of freedom in order to keep the user experience as simple as possible. The data format that we use for describing templates and corresponding annotations is the native format of the semantic web (RDF triples), and we demonstrate some of the ways that generated content can be meaningfully queried using the SPARQL language. We have made all of these materials available as open source (http://github.com/cdd/bioassay-template), in order to encourage community input and use within diverse projects, including but not limited to our own commercial electronic lab notebook products.
Bing
BioAssay templates for the semantic web
Annotation of bioassay protocols using semantic web vocabulary is a way to make experiment descriptions machine-readable. Protocols are communicated using concise scientific English, which precludes most kinds of analysis by software algorithms. Given the availability of a sufficiently expressive ontology, some or all of the pertinent information can be captured by asserting a series of facts, expressed as semantic web triples (subject, predicate, object). With appropriate annotation, assays can be searched, clustered, tagged and evaluated in a multitude of ways, analogous to other segments of drug discovery informatics. The BioAssay Ontology (BAO) has been previously designed for this express purpose, and provides a layered hierarchy of meaningful terms which can be linked to. Currently the biggest challenge is the issue of content creation: scientists cannot be expected to use the BAO effectively without having access to software tools that make it straightforward to use the vocabulary in a canonical way. We have sought to remove this barrier by: (1) defining a BioAssay Template (BAT) data model; (2) creating a software tool for experts to create or modify templates to suit their needs; and (3) designing a common assay template (CAT) to leverage the most value from the BAO terms. The CAT was carefully assembled by biologists in order to find a balance between the maximum amount of information captured vs. low degrees of freedom in order to keep the user experience as simple as possible. The data format that we use for describing templates and corresponding annotations is the native format of the semantic web (RDF triples), and we demonstrate some of the ways that generated content can be meaningfully queried using the SPARQL language. We have made all of these materials available as open source (http://github.com/cdd/bioassay-template), in order to encourage community input and use within diverse projects, including but not limited to our own commercial electronic lab notebook products.
DuckDuckGo
BioAssay templates for the semantic web
Annotation of bioassay protocols using semantic web vocabulary is a way to make experiment descriptions machine-readable. Protocols are communicated using concise scientific English, which precludes most kinds of analysis by software algorithms. Given the availability of a sufficiently expressive ontology, some or all of the pertinent information can be captured by asserting a series of facts, expressed as semantic web triples (subject, predicate, object). With appropriate annotation, assays can be searched, clustered, tagged and evaluated in a multitude of ways, analogous to other segments of drug discovery informatics. The BioAssay Ontology (BAO) has been previously designed for this express purpose, and provides a layered hierarchy of meaningful terms which can be linked to. Currently the biggest challenge is the issue of content creation: scientists cannot be expected to use the BAO effectively without having access to software tools that make it straightforward to use the vocabulary in a canonical way. We have sought to remove this barrier by: (1) defining a BioAssay Template (BAT) data model; (2) creating a software tool for experts to create or modify templates to suit their needs; and (3) designing a common assay template (CAT) to leverage the most value from the BAO terms. The CAT was carefully assembled by biologists in order to find a balance between the maximum amount of information captured vs. low degrees of freedom in order to keep the user experience as simple as possible. The data format that we use for describing templates and corresponding annotations is the native format of the semantic web (RDF triples), and we demonstrate some of the ways that generated content can be meaningfully queried using the SPARQL language. We have made all of these materials available as open source (http://github.com/cdd/bioassay-template), in order to encourage community input and use within diverse projects, including but not limited to our own commercial electronic lab notebook products.
General Meta Tags
29- titleBioAssay templates for the semantic web [PeerJ]
- charsetutf-8
- citation_titleBioAssay templates for the semantic web
- citation_date2016-05-09
- citation_doi10.7717/peerj-cs.61
Open Graph Meta Tags
5- og:imagehttps://dfzljdn9uc3pi.cloudfront.net/2016/cs-61/1/graphical-abstract.jpg
- og:typearticle
- og:titleBioAssay templates for the semantic web
- og:urlhttps://peerj.com/articles/cs-61
- og:site_namePeerJ Computer Science
Twitter Meta Tags
6- twitter:imagehttps://dfzljdn9uc3pi.cloudfront.net/2016/cs-61/1/graphical-abstract.jpg
- twitter:cardsummary_large_image
- twitter:urlhttps://peerj.com/articles/cs-61
- twitter:site@thePeerJ
- twitter:titleBioAssay templates for the semantic web
Item Prop Meta Tags
1- sameAsinfo:doi/10.7717/peerj-cs.61
Link Tags
37- alternate/articles/cs-61.pdf
- alternate/articles/cs-61.rdf
- alternate/articles/cs-61.json
- alternate/articles/cs-61.xml
- alternate/articles/cs-61.unixref
Links
107- http://bioassayexpress.com
- http://bioassayontology.org
- http://bioassayontology.org/bat
- http://bioassayontology.org/bioassayontology
- http://bioportal.bioontology.org