www.nature.com/articles/npjsba201620
Preview meta tags from the www.nature.com website.
Linked Hostnames
34- 131 links towww.nature.com
- 39 links toscholar.google.com
- 37 links todoi.org
- 11 links toscholar.google.co.uk
- 11 links towww.ncbi.nlm.nih.gov
- 7 links towww.springernature.com
- 6 links tolink.springer.com
- 4 links topartnerships.nature.com
Thumbnail
Search Engine Appearance
MINERVA—a platform for visualization and curation of molecular interaction networks - npj Systems Biology and Applications
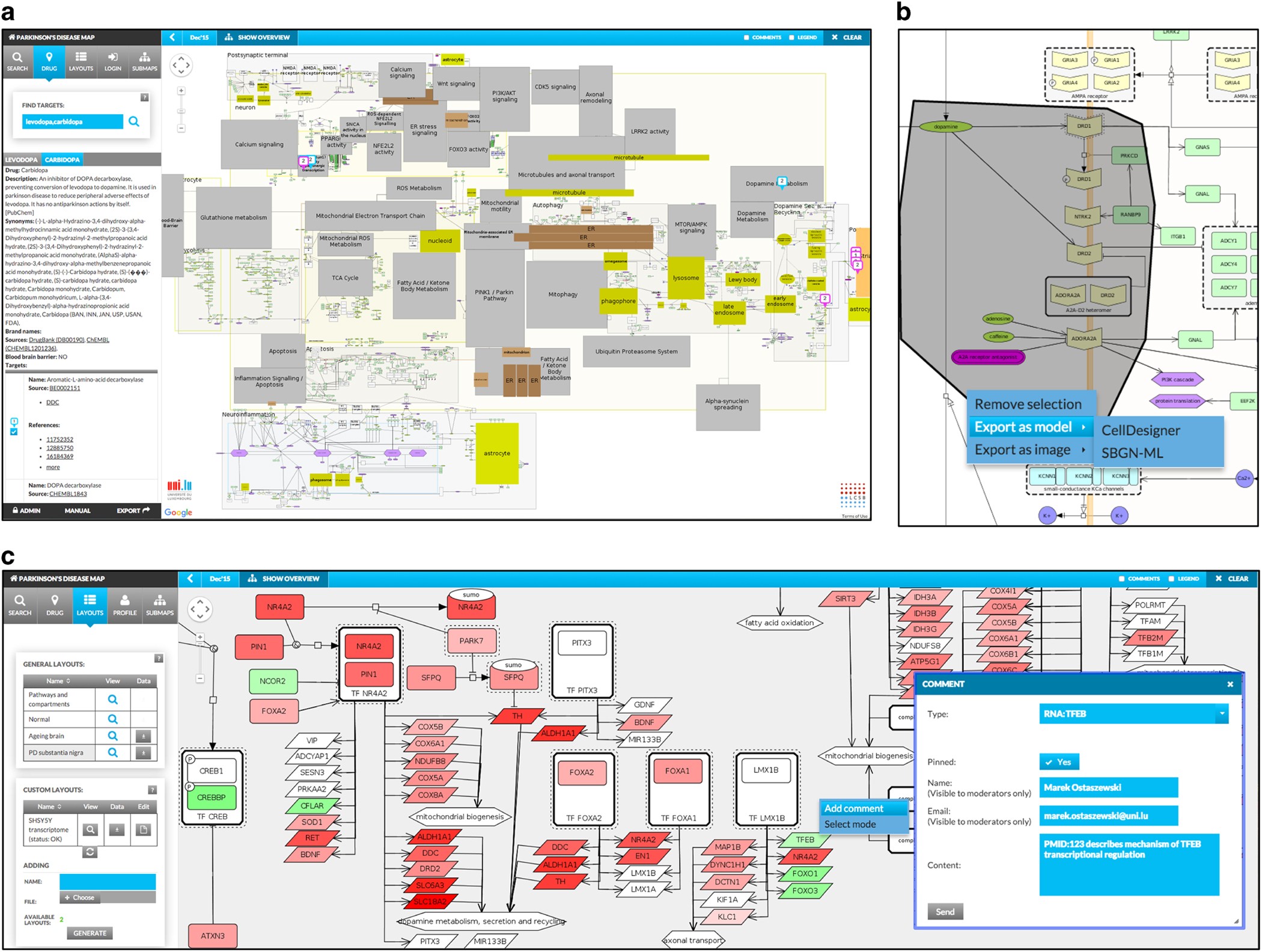
Our growing knowledge about various molecular mechanisms is becoming increasingly more structured and accessible. Different repositories of molecular interactions and available literature enable construction of focused and high-quality molecular interaction networks. Novel tools for curation and exploration of such networks are needed, in order to foster the development of a systems biology environment. In particular, solutions for visualization, annotation and data cross-linking will facilitate usage of network-encoded knowledge in biomedical research. To this end we developed the MINERVA (Molecular Interaction NEtwoRks VisuAlization) platform, a standalone webservice supporting curation, annotation and visualization of molecular interaction networks in Systems Biology Graphical Notation (SBGN)-compliant format. MINERVA provides automated content annotation and verification for improved quality control. The end users can explore and interact with hosted networks, and provide direct feedback to content curators. MINERVA enables mapping drug targets or overlaying experimental data on the visualized networks. Extensive export functions enable downloading areas of the visualized networks as SBGN-compliant models for efficient reuse of hosted networks. The software is available under Affero GPL 3.0 as a Virtual Machine snapshot, Debian package and Docker instance at http://r3lab.uni.lu/web/minerva-website/ . We believe that MINERVA is an important contribution to systems biology community, as its architecture enables set-up of locally or globally accessible SBGN-oriented repositories of molecular interaction networks. Its functionalities allow overlay of multiple information layers, facilitating exploration of content and interpretation of data. Moreover, annotation and verification workflows of MINERVA improve the efficiency of curation of networks, allowing life-science researchers to better engage in development and use of biomedical knowledge repositories. A web platform that integrates data on biological networks can help scientists obtain deeper insights into the inner workings of the cell. Life is powered by the complex interplay between myriad 'pathways' composed of interacting proteins, DNA and other biomolecules. Biologists have accumulated vast troves of data on these processes, and now researchers led by Piotr Gawron at the University of Luxembourg have built software that can combine these disparate datasets into a more meaningful 'big picture'. The Molecular Interaction Networks Visualization (MINERVA) platform allows users to interact directly with graphical representations of biological pathways as a means to understand the broader cellular impacts of both diseases and the drugs intended to treat them. The resulting maps can then be manually curated, annotated and exported to other bioinformatics platforms for use by the broader research community.
Bing
MINERVA—a platform for visualization and curation of molecular interaction networks - npj Systems Biology and Applications
Our growing knowledge about various molecular mechanisms is becoming increasingly more structured and accessible. Different repositories of molecular interactions and available literature enable construction of focused and high-quality molecular interaction networks. Novel tools for curation and exploration of such networks are needed, in order to foster the development of a systems biology environment. In particular, solutions for visualization, annotation and data cross-linking will facilitate usage of network-encoded knowledge in biomedical research. To this end we developed the MINERVA (Molecular Interaction NEtwoRks VisuAlization) platform, a standalone webservice supporting curation, annotation and visualization of molecular interaction networks in Systems Biology Graphical Notation (SBGN)-compliant format. MINERVA provides automated content annotation and verification for improved quality control. The end users can explore and interact with hosted networks, and provide direct feedback to content curators. MINERVA enables mapping drug targets or overlaying experimental data on the visualized networks. Extensive export functions enable downloading areas of the visualized networks as SBGN-compliant models for efficient reuse of hosted networks. The software is available under Affero GPL 3.0 as a Virtual Machine snapshot, Debian package and Docker instance at http://r3lab.uni.lu/web/minerva-website/ . We believe that MINERVA is an important contribution to systems biology community, as its architecture enables set-up of locally or globally accessible SBGN-oriented repositories of molecular interaction networks. Its functionalities allow overlay of multiple information layers, facilitating exploration of content and interpretation of data. Moreover, annotation and verification workflows of MINERVA improve the efficiency of curation of networks, allowing life-science researchers to better engage in development and use of biomedical knowledge repositories. A web platform that integrates data on biological networks can help scientists obtain deeper insights into the inner workings of the cell. Life is powered by the complex interplay between myriad 'pathways' composed of interacting proteins, DNA and other biomolecules. Biologists have accumulated vast troves of data on these processes, and now researchers led by Piotr Gawron at the University of Luxembourg have built software that can combine these disparate datasets into a more meaningful 'big picture'. The Molecular Interaction Networks Visualization (MINERVA) platform allows users to interact directly with graphical representations of biological pathways as a means to understand the broader cellular impacts of both diseases and the drugs intended to treat them. The resulting maps can then be manually curated, annotated and exported to other bioinformatics platforms for use by the broader research community.
DuckDuckGo
MINERVA—a platform for visualization and curation of molecular interaction networks - npj Systems Biology and Applications
Our growing knowledge about various molecular mechanisms is becoming increasingly more structured and accessible. Different repositories of molecular interactions and available literature enable construction of focused and high-quality molecular interaction networks. Novel tools for curation and exploration of such networks are needed, in order to foster the development of a systems biology environment. In particular, solutions for visualization, annotation and data cross-linking will facilitate usage of network-encoded knowledge in biomedical research. To this end we developed the MINERVA (Molecular Interaction NEtwoRks VisuAlization) platform, a standalone webservice supporting curation, annotation and visualization of molecular interaction networks in Systems Biology Graphical Notation (SBGN)-compliant format. MINERVA provides automated content annotation and verification for improved quality control. The end users can explore and interact with hosted networks, and provide direct feedback to content curators. MINERVA enables mapping drug targets or overlaying experimental data on the visualized networks. Extensive export functions enable downloading areas of the visualized networks as SBGN-compliant models for efficient reuse of hosted networks. The software is available under Affero GPL 3.0 as a Virtual Machine snapshot, Debian package and Docker instance at http://r3lab.uni.lu/web/minerva-website/ . We believe that MINERVA is an important contribution to systems biology community, as its architecture enables set-up of locally or globally accessible SBGN-oriented repositories of molecular interaction networks. Its functionalities allow overlay of multiple information layers, facilitating exploration of content and interpretation of data. Moreover, annotation and verification workflows of MINERVA improve the efficiency of curation of networks, allowing life-science researchers to better engage in development and use of biomedical knowledge repositories. A web platform that integrates data on biological networks can help scientists obtain deeper insights into the inner workings of the cell. Life is powered by the complex interplay between myriad 'pathways' composed of interacting proteins, DNA and other biomolecules. Biologists have accumulated vast troves of data on these processes, and now researchers led by Piotr Gawron at the University of Luxembourg have built software that can combine these disparate datasets into a more meaningful 'big picture'. The Molecular Interaction Networks Visualization (MINERVA) platform allows users to interact directly with graphical representations of biological pathways as a means to understand the broader cellular impacts of both diseases and the drugs intended to treat them. The resulting maps can then be manually curated, annotated and exported to other bioinformatics platforms for use by the broader research community.
General Meta Tags
137- titleMINERVA—a platform for visualization and curation of molecular interaction networks | npj Systems Biology and Applications
- titleClose banner
- titleClose banner
- X-UA-CompatibleIE=edge
- applicable-devicepc,mobile
Open Graph Meta Tags
6- og:urlhttps://www.nature.com/articles/npjsba201620
- og:typearticle
- og:site_nameNature
- og:titleMINERVA—a platform for visualization and curation of molecular interaction networks - npj Systems Biology and Applications
- og:descriptionA web platform that integrates data on biological networks can help scientists obtain deeper insights into the inner workings of the cell. Life is powered by the complex interplay between myriad 'pathways' composed of interacting proteins, DNA and other biomolecules. Biologists have accumulated vast troves of data on these processes, and now researchers led by Piotr Gawron at the University of Luxembourg have built software that can combine these disparate datasets into a more meaningful 'big picture'. The Molecular Interaction Networks Visualization (MINERVA) platform allows users to interact directly with graphical representations of biological pathways as a means to understand the broader cellular impacts of both diseases and the drugs intended to treat them. The resulting maps can then be manually curated, annotated and exported to other bioinformatics platforms for use by the broader research community.
Twitter Meta Tags
6- twitter:site@Nature_NPJ
- twitter:cardsummary_large_image
- twitter:image:altContent cover image
- twitter:titleMINERVA—a platform for visualization and curation of molecular interaction networks
- twitter:descriptionnpj Systems Biology and Applications - A web platform that integrates data on biological networks can help scientists obtain deeper insights into the inner workings of the cell. Life is powered by...
Item Prop Meta Tags
5- position1
- position2
- position3
- position4
- publisherSpringer Nature
Link Tags
15- alternatehttps://www.nature.com/npjsba.rss
- apple-touch-icon/static/images/favicons/nature/apple-touch-icon-f39cb19454.png
- canonicalhttps://www.nature.com/articles/npjsba201620
- icon/static/images/favicons/nature/favicon-48x48-b52890008c.png
- icon/static/images/favicons/nature/favicon-32x32-3fe59ece92.png
Emails
1Links
275- http://biocompendium.embl.de
- http://creativecommons.org/licenses/by/4.0
- http://pdmap.uni.lu
- http://r3lab.uni.lu/web/minerva-website
- http://sbgn-ed.org